MSEP: A Platform for Molecular Systems Engineering
MSEP is a free, open-source platform for designing and simulating atomically precise nanomechanical systems — a tool for exploring the foundations of future physical technologies.
The nanoscale mechanism above exists only in simulation today: each sphere represents an atom, and current chemistry can’t guide reactions to form the necessary bonds. Dense, precisely patterned covalent structures like this can’t yet be made, but you can design them now and study how they would work.
Why develop tools for designing molecular machines that we can't yet build? When AI crosses critical capability thresholds, barriers to development will fall, and machines built on these principles will transform the foundations of physical technology. Understanding these prospects is important, and understanding requires concrete exploration.
MSEP: Exploring the World of Molecular Machines
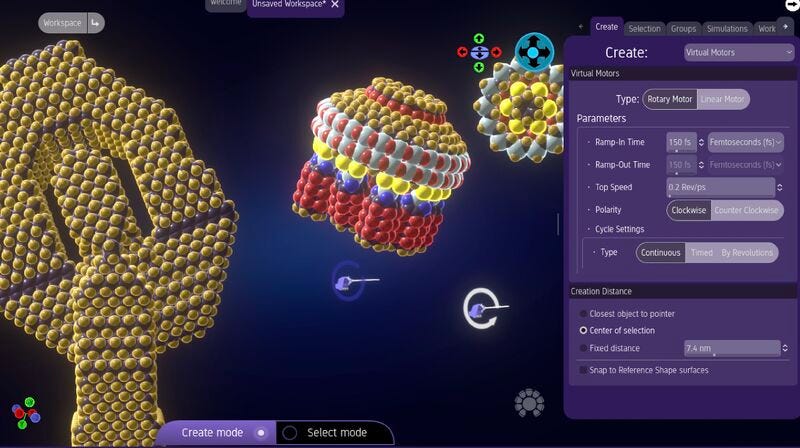
Today marks the public launch of v1.0 of MSEP, the Molecular Systems Engineering Platform. MSEP provides what has been missing: extensible, open-source tools for exploring the space of molecular machines through direct graphical manipulation and simulation. Think Minecraft for molecules, but with physics constraints and atomic precision.
MSEP makes molecular machinery a concrete, explorable domain. Users design mechanical systems, assembling structures and observing their behavior under realistic molecular dynamics.1 Platform extensions will grow the scope of both design and simulation.
Computational tools for molecular simulation have matured over decades, but simulation and intuitive design capabilities rarely intersect. Scientists, understandably, study what exists or can be made in the laboratory, but physical models have no such constraint. It’s only design that calls for different tools. The challenge is to create an environment that combines physics tools with design tools, and to make the software easy to install and easy to use — download, launch, and click to build.2
Built on a modern game engine, MSEP delivers the responsiveness and visual quality users expect from contemporary software, integrated with a modern, extensible architecture that treats physics engines as plug-ins. The platform currently provides atomistic molecular dynamics simulation, with planned extensions including open-source quantum chemistry and machine-oriented multi-scale modeling.
Full disclosure: I’ve helped to support and steer the MSEP development project.
Strategic Leverage Through Early Design
The path from today's protein engineering to self-assembled molecular 3D printer mechanisms (frameworks, moving parts, stepper motors) is clear in outline. This massively parallel molecular positioning capabilitity opens many paths forward forward, though none have yet been described in detail. It is a curious fact that these uncertain paths converge on predictable capabilities that enable atomically precise mass fabrication (APMF). It’s like being able to survey the slopes of mountains in the distance without a map of the landscape nearby.3
When AI systems gain the ability to navigate this path, generative nanotechnologies will be fundamental to the hypercapable world I've described — greatly amplifying AI's impact on physical capabilities. This aligns with the broader trajectory of AI-driven transformation. Advanced AI will expand our general implementation capacity — the ability to design, develop, and deploy complex systems at scale. Molecular systems represent a critical domain where design exploration today can illuminate the landscape of tomorrow's possibilities.
The feedback dynamics deserve emphasis: AI model training requires computational resources that currently consume enormous energy, but atomically precise fabrication will enable post-lithographic devices with million-fold improvements in energy efficiency. When these capabilities converge, the acceleration will be dramatic.
An Invitation to Build
MSEP is available today for Mac, Windows, and Linux at MSEP.one. Download it, explore the physics and — when you’ve developed the skills — share what you create. Fair warning: designing molecular mechanisms combines creative imagination with intricate puzzle-solving. It can be addictive.
The newly established MSEP Foundation (Dutch: Stichting MSEP) will coordinate development and community growth. For those who recognize the strategic importance of building understanding before capabilities arrive, I invite you to support this effort.
» Donate here
Professional computational chemists will recognize that novices can easily build structures that silently violate the conditions for model validity. MSEP v1.0 includes some basic checks, and an integrated Python environment will support the development of additional guardrails and other forms of support for new users.
Point-and-click assembly is an effective way to construct atomic structures for computational models, but real-world fabrication will require sequences of guided encounters between reactive molecules.
Densely bonded covalent structures are extraordinarily stable and readily modeled using molecular dynamics methods, yet intricate architectures of this kind are among the most difficult (today, impossible) to synthesize by conventional chemical means. Their fabrication will require mechanically constrained chemical operations, themselves implemented by post-biomolecular machinery. At every stage, design is the central challenge — one that advanced AI will eventually meet.